Last updated on March 19, 2024 pm
安装补充包
获取非负表达矩阵
使用《使用metacell进行分群聚类》中的数据
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| sce <- readRDS('SRX8890106.rds')
sce@meta.data <- readRDS('SRX8890106_meta.rds')
sce <- subset(sce, seurat_clusters == 6
& DF.classifications_0.25_0.04_416 == 'Singlet')
sce <- Seurat::SCTransform(sce, vst.flavor = "v2", assay = 'RNA',
vars.to.regress = c("CC.Difference", "percent.mt", "percent.rp"),
verbose = F)
DefaultAssay(sce) <- 'RNA'
sce <- Seurat::NormalizeData(sce)
sce <- Seurat::ScaleData(sce, do.center = F,
features = Seurat::VariableFeatures(sce, assay = 'SCT'))
vm <- sce[[Seurat::DefaultAssay(sce)]]@scale.data
|
NMF分解聚类
1
2
3
4
5
6
| saveRDS(vm, 'vm.rds')
vm <- readRDS('vm.rds')
require(NMF)
res <- NMF::nmf(vm, 2:7, method = "snmf/r", seed='ica')
plot(res)
|
1
2
3
4
5
6
7
8
9
10
11
12
13
| require(NMF)
res <- NMF::nmf(vm, 4, method = "snmf/r", seed = 'ica')
DefaultAssay(sce) <- 'SCT'
sce <- Seurat::RunPCA(sce, assay="SCT", verbose = FALSE)
sce@reductions$nmf <- sce@reductions$pca
sce@reductions$nmf@cell.embeddings <- t(coef(res))
sce@reductions$nmf@feature.loadings <- basis(res)
sce <- RunUMAP(sce, reduction = 'nmf', dims = 1:4)
group <- predict(res)
sce$nmf_group <- group[colnames(sce)]
options(repr.plot.width = 6, repr.plot.height = 6)
DimPlot(sce, reduction = "umap", label = T, repel = T,
group.by = c('nmf_group'))
|
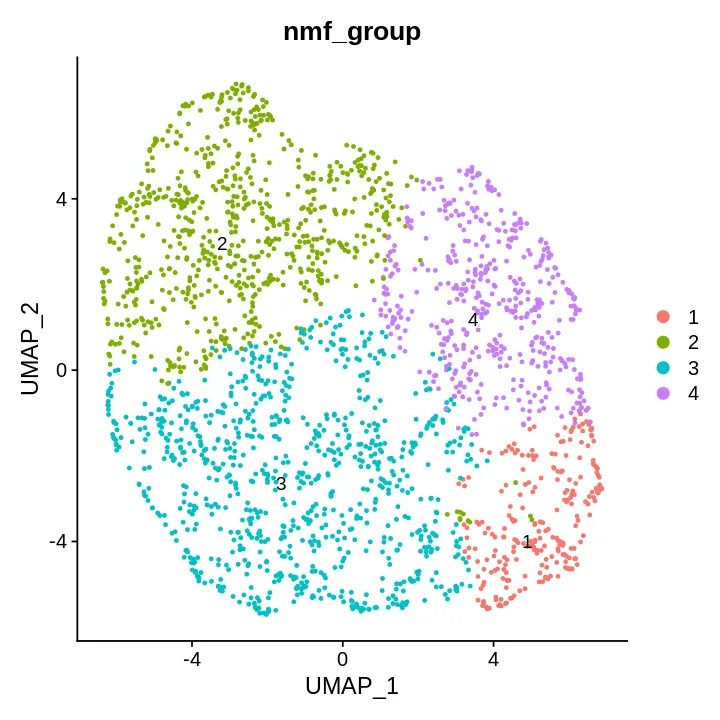
提取signatures
1
2
3
4
5
6
| coefmap(res)
consensusmap(res)
df <- extractFeatures(res, 20L)
df <- lapply(df, function(x){rownames(res)[x]})
df <- as.data.frame(do.call("rbind", df))
df
|
【迁移】基于NMF分解的降维聚类
https://hexo.limour.top/ji-yu-NMF-fen-jie-de-jiang-wei-ju-lei
