1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
|
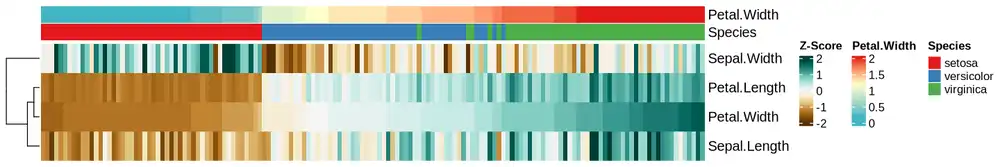
col_Petal.Width <- colorRamp2(
c(0.2, 1.2, 2),
c(
RColorBrewer::brewer.pal(9, 'YlGnBu')[5],
RColorBrewer::brewer.pal(9, 'YlOrRd')[1],
RColorBrewer::brewer.pal(9, 'YlOrRd')[7]
)
)
col_ANO = list(
Species = c(
"setosa" = RColorBrewer::brewer.pal(9, 'Set1')[1],
"versicolor" = RColorBrewer::brewer.pal(9, 'Set1')[2],
"virginica" = RColorBrewer::brewer.pal(9, 'Set1')[3]
),
Petal.Width = col_Petal.Width
)
ha <- HeatmapAnnotation(
Petal.Width = data$Petal.Width,
Species = data$Species,
col = col_ANO,
show_annotation_name = TRUE
)
|